Method bbm
bbm.RdFunction to fit a two-stage biomass-based model.
bbm(object, indicesB, indicesP, ...)
# S4 method for FLQuant,FLQuants,FLQuants
bbm(object, indicesB, indicesP, findicesB, findicesP, control, inits)
# S4 method for FLStock,ANY,ANY
bbm(
object,
indicesB,
indicesP,
findicesB = NULL,
findicesP = NULL,
control,
inits
)
# S4 method for FLQuant,FLIndices,FLIndices
bbm(
object,
indicesB,
indicesP,
findicesB = NULL,
findicesP = NULL,
control,
inits
)
# S4 method for FLQuant,FLQuant,FLQuant
bbm(object, indicesB, indicesP, findicesB, findicesP, control, inits)Arguments
- object
An
FLQuantwith catch information (in mass) or anFLStock. These objects must only have two age classes (recruits and adults) and the number of seasons should be 1 or the number of seasons determined by the timing of the different indices.- indicesB
Abundance indices in total biomass (element of class:
FLQuants,FLQuantorFLIndices) from surveys. Please assign a survey name to each index.- indicesP
Percentage of recruits in biomass (element of class:
FLQuants,FLQuantorFLIndices) from surveys. Please assign a survey name to each proportion.- findicesB
A
vectorwith fraction of the year corresponding to each of the indicesB.- findicesP
A
vectorwith fraction of the year corresponding to each of the indicesP.- control
A
bbmControlwith control arguments.- inits
An
FLParwith initial values.
Value
An object of class bbmFit.
See also
bbmFit, FLQuant, FLQuants, bbmControl, FLPar, bbmFLPar
Examples
# Load data
data(ane)
# Case: object='FLStock'; indicesB=indicesP='FLIndices'; control='bbmControl'; inits='FLPar'
stock <- FLStock(catch.n=catch.ane, catch.wt=catch.ane*0+1)
units(stock@catch.wt) <- ''
stock@catch <- quantSums(stock@catch.n*stock@catch.wt)
run2 <- bbm(stock, indicesB=indicesB.ane, indicesP=indicesP.ane, control=control.ane, inits=inits.ane)
#> Constructing atomic D_lgamma
#> outer mgc: 2.991218
#> outer mgc: 2.983554
#> outer mgc: 2.994433
#> outer mgc: 2.980339
#> outer mgc: 2.989589
#> outer mgc: 2.985185
#> outer mgc: 2.990111
#> outer mgc: 2.984663
#> outer mgc: 2.986485
#> outer mgc: 2.988286
#> outer mgc: 2.987929
#> outer mgc: 2.986843
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987385
#> outer mgc: 2.987387
#> outer mgc: 2.987385
#> outer mgc: 2.987387
#> outer mgc: 2.987382
#> outer mgc: 2.98739
#> outer mgc: 2.987383
#> outer mgc: 2.987388
#> outer mgc: 2.98738
#> outer mgc: 2.987392
#> outer mgc: 2.987369
#> outer mgc: 2.987403
#> outer mgc: 2.987363
#> outer mgc: 2.987409
#> outer mgc: 2.987322
#> outer mgc: 2.98745
#> outer mgc: 2.98723
#> outer mgc: 2.987541
#> outer mgc: 2.987124
#> outer mgc: 2.987647
#> outer mgc: 2.986773
#> outer mgc: 2.987998
#> outer mgc: 2.986082
#> outer mgc: 2.988689
#> outer mgc: 2.986608
#> outer mgc: 2.988164
#> outer mgc: 3.008474
#> outer mgc: 2.96631
#> outer mgc: 2.986905
#> outer mgc: 2.987863
#> outer mgc: 2.986374
#> outer mgc: 2.988397
#> outer mgc: 2.986725
#> outer mgc: 2.988047
#> outer mgc: 2.9823
#> outer mgc: 2.992471
#> outer mgc: 2.985849
#> outer mgc: 2.988921
#> outer mgc: 2.987386
#> outer mgc: 2.991218
#> outer mgc: 2.983554
#> outer mgc: 2.994433
#> outer mgc: 2.980339
#> outer mgc: 2.989589
#> outer mgc: 2.985185
#> outer mgc: 2.990111
#> outer mgc: 2.984663
#> outer mgc: 2.986485
#> outer mgc: 2.988286
#> outer mgc: 2.987929
#> outer mgc: 2.986843
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987385
#> outer mgc: 2.987387
#> outer mgc: 2.987385
#> outer mgc: 2.987387
#> outer mgc: 2.987382
#> outer mgc: 2.98739
#> outer mgc: 2.987383
#> outer mgc: 2.987388
#> outer mgc: 2.98738
#> outer mgc: 2.987392
#> outer mgc: 2.987369
#> outer mgc: 2.987403
#> outer mgc: 2.987363
#> outer mgc: 2.987409
#> outer mgc: 2.987322
#> outer mgc: 2.98745
#> outer mgc: 2.98723
#> outer mgc: 2.987541
#> outer mgc: 2.987124
#> outer mgc: 2.987647
#> outer mgc: 2.986773
#> outer mgc: 2.987998
#> outer mgc: 2.986082
#> outer mgc: 2.988689
#> outer mgc: 2.986608
#> outer mgc: 2.988164
#> outer mgc: 3.008474
#> outer mgc: 2.96631
#> outer mgc: 2.986905
#> outer mgc: 2.987863
#> outer mgc: 2.986374
#> outer mgc: 2.988397
#> outer mgc: 2.986725
#> outer mgc: 2.988047
#> outer mgc: 2.9823
#> outer mgc: 2.992471
#> outer mgc: 2.985849
#> outer mgc: 2.988921
#> outer mgc: 282597.9
#> Warning: NaNs produced
#> Warning: NaNs produced
# Case: object='FLQuant'; indicesB=indicesP='FLIndices'; control='bbmControl'; inits='FLPar'
run3 <- bbm(catch.ane, indicesB=indicesB.ane, indicesP=indicesP.ane, control=control.ane, inits=inits.ane)
#> outer mgc: 2.991218
#> outer mgc: 2.983554
#> outer mgc: 2.994433
#> outer mgc: 2.980339
#> outer mgc: 2.989589
#> outer mgc: 2.985185
#> outer mgc: 2.990111
#> outer mgc: 2.984663
#> outer mgc: 2.986485
#> outer mgc: 2.988286
#> outer mgc: 2.987929
#> outer mgc: 2.986843
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987385
#> outer mgc: 2.987387
#> outer mgc: 2.987385
#> outer mgc: 2.987387
#> outer mgc: 2.987382
#> outer mgc: 2.98739
#> outer mgc: 2.987383
#> outer mgc: 2.987388
#> outer mgc: 2.98738
#> outer mgc: 2.987392
#> outer mgc: 2.987369
#> outer mgc: 2.987403
#> outer mgc: 2.987363
#> outer mgc: 2.987409
#> outer mgc: 2.987322
#> outer mgc: 2.98745
#> outer mgc: 2.98723
#> outer mgc: 2.987541
#> outer mgc: 2.987124
#> outer mgc: 2.987647
#> outer mgc: 2.986773
#> outer mgc: 2.987998
#> outer mgc: 2.986082
#> outer mgc: 2.988689
#> outer mgc: 2.986608
#> outer mgc: 2.988164
#> outer mgc: 3.008474
#> outer mgc: 2.96631
#> outer mgc: 2.986905
#> outer mgc: 2.987863
#> outer mgc: 2.986374
#> outer mgc: 2.988397
#> outer mgc: 2.986725
#> outer mgc: 2.988047
#> outer mgc: 2.9823
#> outer mgc: 2.992471
#> outer mgc: 2.985849
#> outer mgc: 2.988921
#> outer mgc: 2.987386
#> outer mgc: 2.991218
#> outer mgc: 2.983554
#> outer mgc: 2.994433
#> outer mgc: 2.980339
#> outer mgc: 2.989589
#> outer mgc: 2.985185
#> outer mgc: 2.990111
#> outer mgc: 2.984663
#> outer mgc: 2.986485
#> outer mgc: 2.988286
#> outer mgc: 2.987929
#> outer mgc: 2.986843
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987386
#> outer mgc: 2.987385
#> outer mgc: 2.987387
#> outer mgc: 2.987385
#> outer mgc: 2.987387
#> outer mgc: 2.987382
#> outer mgc: 2.98739
#> outer mgc: 2.987383
#> outer mgc: 2.987388
#> outer mgc: 2.98738
#> outer mgc: 2.987392
#> outer mgc: 2.987369
#> outer mgc: 2.987403
#> outer mgc: 2.987363
#> outer mgc: 2.987409
#> outer mgc: 2.987322
#> outer mgc: 2.98745
#> outer mgc: 2.98723
#> outer mgc: 2.987541
#> outer mgc: 2.987124
#> outer mgc: 2.987647
#> outer mgc: 2.986773
#> outer mgc: 2.987998
#> outer mgc: 2.986082
#> outer mgc: 2.988689
#> outer mgc: 2.986608
#> outer mgc: 2.988164
#> outer mgc: 3.008474
#> outer mgc: 2.96631
#> outer mgc: 2.986905
#> outer mgc: 2.987863
#> outer mgc: 2.986374
#> outer mgc: 2.988397
#> outer mgc: 2.986725
#> outer mgc: 2.988047
#> outer mgc: 2.9823
#> outer mgc: 2.992471
#> outer mgc: 2.985849
#> outer mgc: 2.988921
#> outer mgc: 282597.9
#> Warning: NaNs produced
#> Warning: NaNs produced
# Case: object='FLQuant'; indicesB=indicesP='FLQuant'; control='bbmControl'; inits='FLPar'
namdel <- c("q_acoustic","psi_acoustic","xi_acoustic") # we will take only one of the indices --> need to delete the parameters related to other indices
control <- control.ane
control@param.fix <- control@param.fix[dimnames(control@param.fix)$params[!dimnames(control@param.fix)$params %in% namdel],]
inits <- inits.ane[dimnames(inits.ane)$params[!dimnames(inits.ane)$params %in% namdel],]
run4 <- bbm( catch.ane, indicesB=indicesB.ane[[1]]@index, indicesP=indicesP.ane[[1]]@index,
findicesB=c( depm=(indicesB.ane[[1]]@range[['startf']]+indicesB.ane[[1]]@range[['endf']])/2),
findicesP=c( depm=(indicesP.ane[[1]]@range[['startf']]+indicesP.ane[[1]]@range[['endf']])/2),
control=control, inits=inits)
#> outer mgc: 8.017707
#> outer mgc: 8.020517
#> outer mgc: 8.020002
#> outer mgc: 8.018223
#> outer mgc: 8.021608
#> outer mgc: 8.01662
#> outer mgc: 8.019126
#> outer mgc: 8.019099
#> outer mgc: 8.019138
#> outer mgc: 8.019086
#> outer mgc: 8.019161
#> outer mgc: 8.019064
#> outer mgc: 8.019206
#> outer mgc: 8.019019
#> outer mgc: 8.019293
#> outer mgc: 8.018932
#> outer mgc: 8.019458
#> outer mgc: 8.018766
#> outer mgc: 7.937066
#> outer mgc: 8.101158
#> outer mgc: 8.016787
#> outer mgc: 8.021442
#> outer mgc: 8.018655
#> outer mgc: 8.01957
#> outer mgc: 8.019304
#> outer mgc: 8.018921
#> outer mgc: 8.01896
#> outer mgc: 8.019265
#> outer mgc: 8.019135
#> outer mgc: 8.01909
#> outer mgc: 8.019125
#> outer mgc: 8.0191
#> outer mgc: 8.019095
#> outer mgc: 8.01913
#> outer mgc: 8.019094
#> outer mgc: 8.01913
#> outer mgc: 8.019103
#> outer mgc: 8.019121
#> outer mgc: 8.019111
#> outer mgc: 8.019114
#> outer mgc: 8.019113
#> outer mgc: 8.019112
#> outer mgc: 8.019113
#> outer mgc: 8.019112
#> outer mgc: 8.019113
#> outer mgc: 8.019112
#> outer mgc: 8.019112
#> outer mgc: 8.019112
#> outer mgc: 8.097982
#> outer mgc: 7.940243
#> outer mgc: 8.023486
#> outer mgc: 8.014743
#> outer mgc: 8.019112
#> outer mgc: 8.017707
#> outer mgc: 8.020517
#> outer mgc: 8.020002
#> outer mgc: 8.018223
#> outer mgc: 8.021608
#> outer mgc: 8.01662
#> outer mgc: 8.019126
#> outer mgc: 8.019099
#> outer mgc: 8.019138
#> outer mgc: 8.019086
#> outer mgc: 8.019161
#> outer mgc: 8.019064
#> outer mgc: 8.019206
#> outer mgc: 8.019019
#> outer mgc: 8.019293
#> outer mgc: 8.018932
#> outer mgc: 8.019458
#> outer mgc: 8.018766
#> outer mgc: 7.937066
#> outer mgc: 8.101158
#> outer mgc: 8.016787
#> outer mgc: 8.021442
#> outer mgc: 8.018655
#> outer mgc: 8.01957
#> outer mgc: 8.019304
#> outer mgc: 8.018921
#> outer mgc: 8.01896
#> outer mgc: 8.019265
#> outer mgc: 8.019135
#> outer mgc: 8.01909
#> outer mgc: 8.019125
#> outer mgc: 8.0191
#> outer mgc: 8.019095
#> outer mgc: 8.01913
#> outer mgc: 8.019094
#> outer mgc: 8.01913
#> outer mgc: 8.019103
#> outer mgc: 8.019121
#> outer mgc: 8.019111
#> outer mgc: 8.019114
#> outer mgc: 8.019113
#> outer mgc: 8.019112
#> outer mgc: 8.019113
#> outer mgc: 8.019112
#> outer mgc: 8.019113
#> outer mgc: 8.019112
#> outer mgc: 8.019112
#> outer mgc: 8.019112
#> outer mgc: 8.097982
#> outer mgc: 7.940243
#> outer mgc: 8.023486
#> outer mgc: 8.014743
#> outer mgc: 94064.07
#> Warning: NaNs produced
#> Warning: NaNs produced
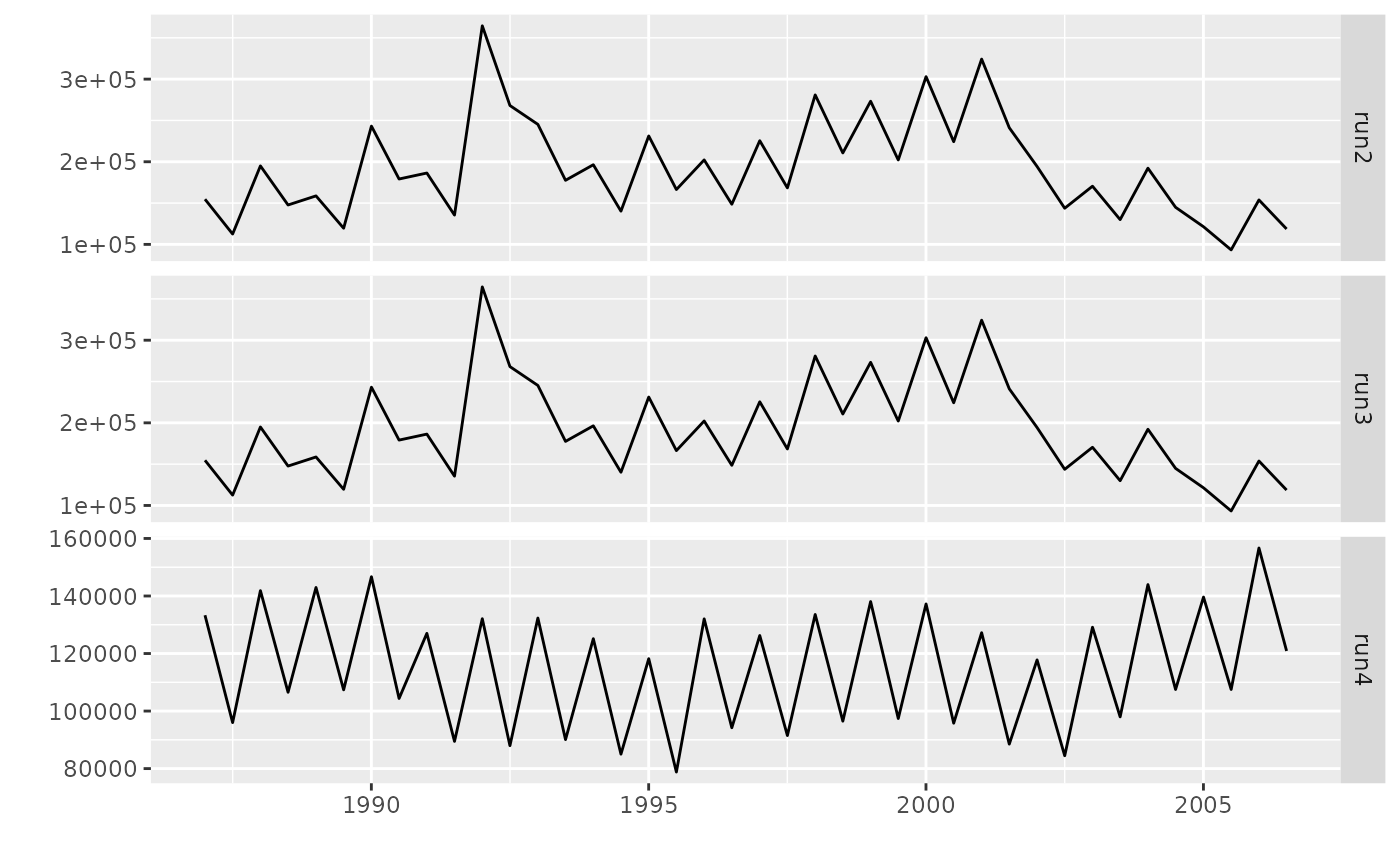
# Plot assessed populations
runs <- list(run2=run2, run3=run3, run4=run4)
biomass <- FLQuants(lapply(lapply(runs, stock.bio), quantSums))
plot(biomass)